
Cytoscape network update#
Gallery Dynamically expand elementsĬode | Demo Interactively update stylesheetĬode | Demo Automatically generate interactive phylogeny treesįor an extended gallery, visit the demos' readme. The Pull Request and Issue Templates were inspired from the This library would not have been possible without their massive work! Huge thanks to the Cytoscape Consortium and the Cytoscape.js team for their contribution in making such a complete API for creating interactive networks. Licenseĭash, Cytoscape.js and Dash Cytoscape are licensed under MIT.
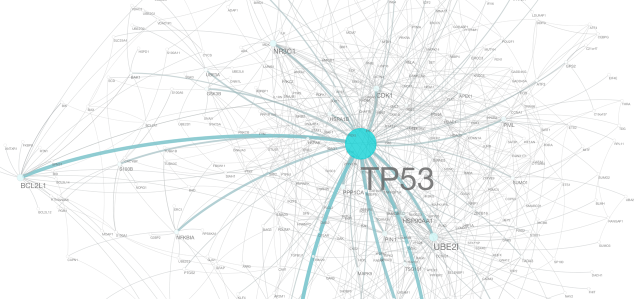
Network diameter, radius and clustering coefficient, as well as the characteristic path length.
Cytoscape network how to#
Instructions on how to run tests are given in CONTRIBUTING.md. NetworkAnalyzer NetworkAnalyzer computes a comprehensive set of topological parameters for undirected and directed networks, including: Number of nodes, edges and connected components. In the situations described above, NetworkAnalyzer needs user input how to interpret the edges. Moreover, one network may contain both directed and undirected edges if the network is compiled by combining data from different sources. Make sure that you have read and understood our code of conduct, then head over to CONTRIBUTING to get started. In Cytoscape, a network may contain only directed edges even if they are undirected in their biological context. To learn more about the core Dash components and how to use callbacks, view the Dash documentation.įor supplementary information about the underlying Javascript API, view the Cytoscape.js documentation. You can also use the component reference for a complete and concise specification of the API. It contains useful examples, functioning code, and is fully interactive. The Dash Cytoscape User Guide contains everything you need to know about the library. A Cytoscape App for multiple local alignment of protein-protein interaction (PPI) networks InsituNet Analysis of in situ gene expression data in terms of spatial co-expression.

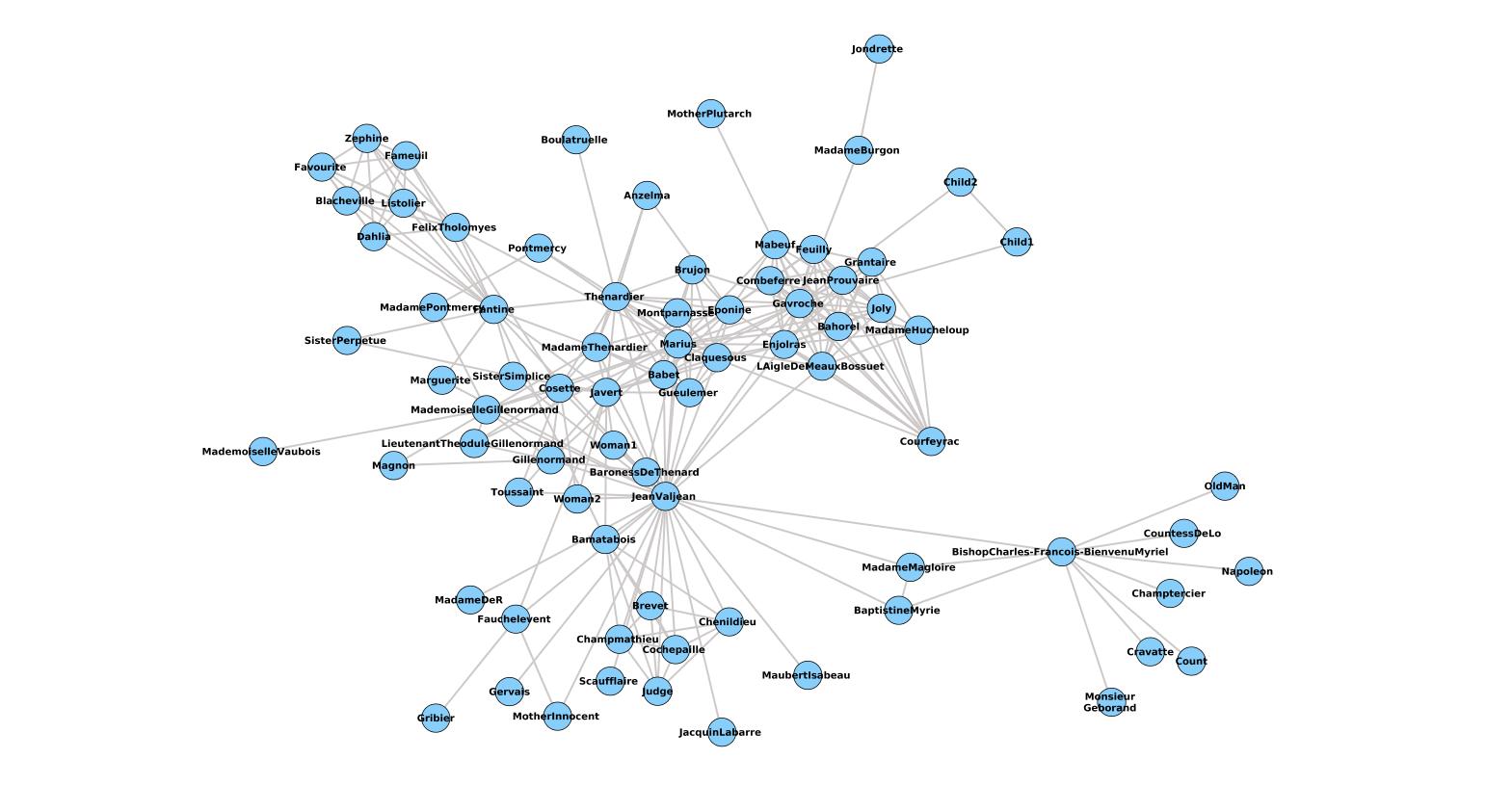
List( 'data ' = list( 'source ' = 'one ', 'target ' = 'two ')) The GUI,depicted inFigure 10.2, provides asimple way touse the plugin: Once a network has been loaded into Cytoscape,users canselect the whole network ora. Style = list( 'width ' = '100% ', 'height ' = '400px '), Cytoscape is a tool for viewing and analyzing networks (meaning, in this case, any group of entities that are connected in some way).


 0 kommentar(er)
0 kommentar(er)
